We’ll also interpret the found linear dependencies. That means we check whether the discovered pattern corresponds to common sense. The main purpose of the task is to show and explain by example what causes overfitting and how to overcome it.
The code as an IPython notebook
Table of Contents
- Feature target plots
- Pairwise correlations between all featues except for the target
- Pairwise correlations between each feature and the target
- Train linear model
- Model coefficients as function of alpha
- The weights dependence on alpha dynamic
- Sub-conclusions on Lasso and Ridge regularizators
- Cross-validation
- Plotting the mean MSE as a function of alpha.
- Sub-conclusion on the Lasso regularization
Features and target correlations
Problem one: Collinear (highly correlated) features
Problem two: Uninformative features
We will work with Lasso (L1) regularizator to optimize regression model with regularization
Dataset
We will use the bikes_rent.csv file as a dataset, which records calendar information and weather conditions that characterize automated bike rental locations and the target: number of rentals on that day, named cnt. The latter we will predict them in linear regression models and evaluate models quality. Thus, we will solve the regression problem.
import pandas as pd import numpy as np from matplotlib import pyplot as plt %matplotlib inline
df = pd.read_csv("bikes_rent.csv")
df.head()
season yr mnth holiday weekday workingday weathersit
0 1 0 1 0 6 0 2
1 1 0 1 0 0 0 2
2 1 0 1 0 1 1 1
3 1 0 1 0 2 1 1
4 1 0 1 0 3 1 1
temp atemp hum windspeed(mph) windspeed(ms) cnt
0 14.110847 18.18125 80.5833 10.749882 4.805490 985
1 14.902598 17.68695 69.6087 16.652113 7.443949 801
2 8.050924 9.47025 43.7273 16.636703 7.437060 1349
3 8.200000 10.60610 59.0435 10.739832 4.800998 1562
4 9.305237 11.46350 43.6957 12.522300 5.597810 1600
For each rental day, the following attributes/dataset features are known (as they were specified in the data source):
- season: 1-spring, 2-summer, 3-autumn, 4-winter
- yr: 0 – 2011, 1 – 2012
- mnth: from 1 to 12
- holiday: 0 – no holiday, 1 – there is a holiday
- weekday: 0 to 6
- workingday: 0 – non-working day, 1 – working day
- weathersit: weather favorability rating from 1 (clear day) to 4 (rain, fog)
- temp: temperature in Celsius degrees
- atemp: temperature feels like, Celsius degrees
- hum: humidity
- windspeed(mph): wind speed in miles per hour
- windspeed (ms): wind speed in meters per second
- cnt: the number of rented bicycles (this is the target attribute, we will predict it thru a built model)
So, we have real (temp atemp, hum, windspeed(mph), windspeed(ms)), binary (holiday, workingday), and nominal (ordinal) features (eg. season: 1-spring, 2-summer, 3-autumn, 4-winter; weekday, weathersit). All of them might be worked with as real features.
One can work with nominal attributes as with real ones, because they are in an order. Let’s look at the figures below, showing how the target attribute (cnt) depends on each of the features.
Feature target plots
fig, axes = plt.subplots(nrows=3, ncols=4, figsize=(15, 10))
for idx, feature in enumerate(df.columns[:-1]):
df.plot(feature, "cnt", subplots=True, kind="scatter", ax=axes[idx // 4, idx % 4])
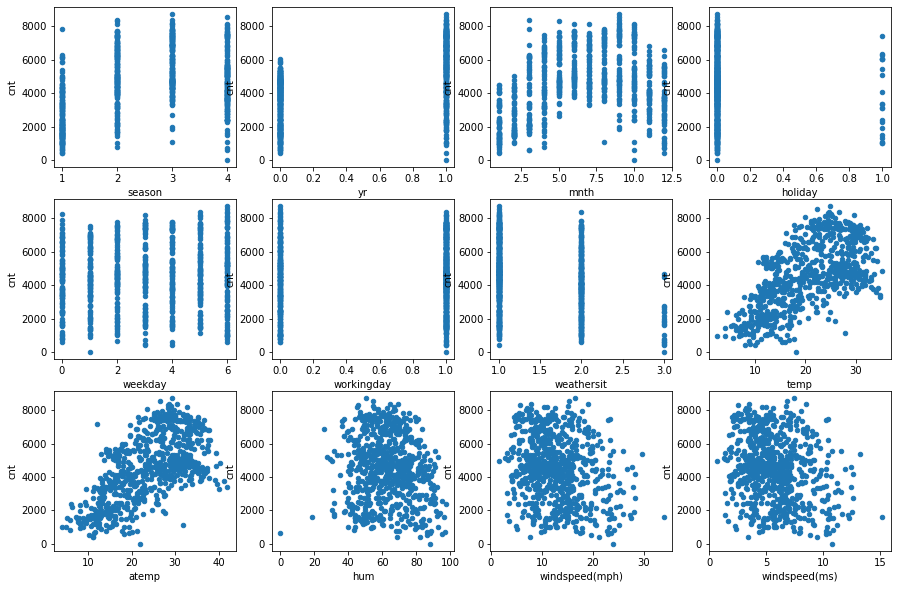
One can see from the charts that there is the linear dependence of the target of some features, eg. season, atemp.
Features and target correlations
Pairwise correlations between all featues except for the target
Let’s more strictly estimate the level of linear dependence between the features and the target variable. A good measure of the linear relationship between two vectors is the Pearson correlation. In the Pandas it can be calculated using two dataframe methods: corr and corrwith. The df.corr method calculates the correlation matrix of all features from the dataframe. The df.corrwith method needs to supply another dataframe as an argument, and then it will calculate pairwise correlations between the features from the function argument and the original dataframe.
# df2 - df without the column "cnt", the target df2 = df[df.columns[:-1]] # alternative: df2 = df[df.columns.difference(["cnt"])] # get features correlation coefficients df2.corr(method="pearson")
season yr mnth holiday weekday workingday weathersit temp atemp hum windspd(mph) windspd(ms) season 1.000000 -0.001844 0.831440 -0.010537 -0.003080 0.012485 0.019211 0.334315 0.342876 0.205445 -0.229046 -0.229046 yr -0.001844 1.000000 -0.001792 0.007954 -0.005461 -0.002013 -0.048727 0.047604 0.046106 -0.110651 -0.011817 -0.011817 mnth 0.831440 -0.001792 1.000000 0.019191 0.009509 -0.005901 0.043528 0.220205 0.227459 0.222204 -0.207502 -0.207502 holiday -0.010537 0.007954 0.019191 1.000000 -0.101960 -0.253023 -0.034627 -0.028556 -0.032507 -0.015937 0.006292 0.006292 weekday -0.003080 -0.005461 0.009509 -0.101960 1.000000 0.035790 0.031087 -0.000170 -0.007537 -0.052232 0.014282 0.014282 workingday 0.012485 -0.002013 -0.005901 -0.253023 0.035790 1.000000 0.061200 0.052660 0.052182 0.024327 -0.018796 -0.018796 weathersit 0.019211 -0.048727 0.043528 -0.034627 0.031087 0.061200 1.000000 -0.120602 -0.121583 0.591045 0.039511 0.039511 temp 0.334315 0.047604 0.220205 -0.028556 -0.000170 0.052660 -0.120602 1.000000 0.991702 0.126963 -0.157944 -0.157944 atemp 0.342876 0.046106 0.227459 -0.032507 -0.007537 0.052182 -0.121583 0.991702 1.000000 0.139988 -0.183643 -0.183643 hum 0.205445 -0.110651 0.222204 -0.015937 -0.052232 0.024327 0.591045 0.126963 0.139988 1.000000 -0.248489 -0.248489 windspeed(mph) -0.229046 -0.011817 -0.207502 0.006292 0.014282 -0.018796 0.039511 -0.157944 -0.183643 -0.248489 1.000000 1.000000 windspeed(ms) -0.229046 -0.011817 -0.207502 0.006292 0.014282 -0.018796 0.039511 -0.157944 -0.183643 -0.248489 1.000000 1.000000
Pairwise correlations between each feature and the target
# kitchen sink solution: pd.concat([df,df],axis=1).corr() df.corrwith(df["cnt"], axis=0, method="pearson")
season 0.406100 yr 0.566710 mnth 0.279977 holiday -0.068348 weekday 0.067443 workingday 0.061156 weathersit -0.297391 temp 0.627494 atemp 0.631066 hum -0.100659 windspeed(mph) -0.234545 windspeed(ms) -0.234545 cnt 1.000000 dtype: float64
The sample above shows features correlations with the target. All of them are not null. That means that we can solve the problem using linear methods.
The graphs above show that some of the features are similar to each other. So let’s also calculate the correlations between the real features: temp, atemp, hum, windspeed(mph), windspeed(ms) and cnt
df3 = df[["temp", "atemp", "hum", "windspeed(mph)", "windspeed(ms)", "cnt"]] df3.corr(method="pearson")
temp atemp hum windspeed(mph) windspeed(ms) cnt temp 1.000000 0.991702 0.126963 -0.157944 -0.157944 0.627494 atemp 0.991702 1.000000 0.139988 -0.183643 -0.183643 0.631066 hum 0.126963 0.139988 1.000000 -0.248489 -0.248489 -0.100659 windspd(mph) -0.157944 -0.183643 -0.248489 1.000000 1.000000 -0.234545 windspeed(ms) -0.157944 -0.183643 -0.248489 1.000000 1.000000 -0.234545 cnt 0.627494 0.631066 -0.100659 -0.234545 -0.234545 1.000000
As expected, there are ones 1 on the diagonals. However, there are two more pairs of strongly correlated columns in the matrix: temp and atemp (correlated by nature) and two windspeed features (because one is just a translation of some units into others). Later we will see that this effects negatively on the training of the linear model.
Let’s take a look at the mean values of the features to estimate the scale of the features and the fraction of 1 for binary features.
df.mean()
season 2.496580 yr 0.500684 mnth 6.519836 holiday 0.028728 weekday 2.997264 workingday 0.683995 weathersit 1.395349 temp 20.310776 atemp 23.717699 hum 62.789406 windspeed(mph) 12.762576 windspeed(ms) 5.705220 cnt 4504.348837 dtype: float64
The features are of different scale. So for our further work, we better normalize the feature objects matrix.
Problem one: Collinear (highly correlated) features
So, in our data, some attributes duplicates another ones and there are two more very similar ones. Of course, we could remove the duplicates right away, but let’s see how model would have been trained if we hadn’t noticed this problem.
To begin with, we will scale or standardize the features. From each feature, we will subtract its average (mean) and divide it by the standard deviation (std). This can be done using the scale() method.
In addition we shuffle the dataset; this is required for cross-validation.
from sklearn.preprocessing import scale from sklearn.utils import shuffle
df_shuffled = shuffle(df, random_state=123) print(df_shuffled.head()) X = scale(df_shuffled[df_shuffled.columns[:-1]]) y = df_shuffled["cnt"]
season yr mnth holiday weekday workingday weathersit temp \
488 2 1 5 0 4 1 2 22.960000
421 1 1 2 0 0 0 1 11.445847
91 2 0 4 0 6 0 2 12.915000
300 4 0 10 0 5 1 2 13.564153
177 3 0 6 0 1 1 2 27.982500
atemp hum windspeed(mph) windspeed(ms) cnt
488 26.86210 76.8333 8.957632 4.004306 6421
421 13.41540 41.0000 13.750343 6.146778 3389
91 15.78185 65.3750 13.208782 5.904686 2252
300 15.94060 58.5833 15.375093 6.873086 3747
177 31.85020 65.8333 7.208396 3.222350 4708
Train linear model
Let’s train a linear regression model on our dataset and look at the feature weights. The acquired weights are in the coef_ parameter of a regressor class.
from sklearn.linear_model import LinearRegression
from sklearn import linear_model
# build a model
linear_regressor = linear_model.LinearRegression()
# train the model using all data
linear_regressor.fit(X, y)
# output coefficients
print("Linear model coefficients:\n", linear_regressor.coef_ )
# get the model predictions
# predictions = linear_regressor.predict()
print("\nCoefficients and their weights:")
[(pair[0], round(pair[1],2)) for pair in zip(df.columns, linear_regressor.coef_)]
Linear model coefficients: [ 5.70865954e+02 1.02196587e+03 -1.41301029e+02 -8.67565864e+01 1.37223034e+02 5.63936928e+01 -3.30228163e+02 3.67479222e+02 5.85554695e+02 -1.45612324e+02 1.24575770e+13 -1.24575770e+13]
Coefficients and their weights:
[("season", 570.87),
("yr", 1021.97),
("mnth", -141.3),
("holiday", -86.76),
("weekday", 137.22),
("workingday", 56.39),
("weathersit", -330.23),
("temp", 367.48),
("atemp", 585.55),
("hum", -145.61),
("windspeed(mph)", 12457576985764.57),
("windspeed(ms)", -12457576985963.02)]
We see that the weights for linearly dependent features are significantly greater in absolute values than for other features.
To better understand why this happened, let’s recall the analytical formula used to calculate the weights of a linear model in the least squares method:
w = (XTX)-1* XT* y
If there are collinear (linearly dependent) columns in X, the matrix XTX becomes degenerate one. The formula then ceases to be correct. The more dependent the features are, the smaller the determinant of this matrix is and the worse the approximation of 𝑋*𝑤≈𝑦 is. This situation is called the multicollinearity problem.
The temp-atemp pair had slightly less correlated variables/weights. But in practice, one should always carefully monitor the coefficients for similar attributes.
The multicollinearity problem solution is to regularize the linear model. The norm of weights multiplied by the regularization coefficient alpha, α, (L1 or L2), is added to the optimized functional X*w. In the first case (L1), the method is called Lasso, and in the second (L2), the method is called Ridge. Read more about the adding regularization into a Linear Regression model.
We train the Ridge и Lasso regressors with the default parameters and make sure that the problem with the weights is resolved.
from sklearn.linear_model import Lasso, Ridge
lasso = Lasso()
lasso.fit(X, y)
print("Lasso regularization coefficients/weights:\n", lasso.coef_)
print("\nIntercept:", lasso.intercept_)
print( "\nRounded coef:", list(map(lambda c: round(c, 3) , lasso.coef_) ))
Lasso regularization coefficients/weights: [ 5.60241616e+02 1.01946349e+03 -1.28730627e+02 -8.61527813e+01 1.37347894e+02 5.52123706e+01 -3.32369857e+02 3.76363236e+02 5.76530794e+02 -1.44129155e+02 -1.97139689e+02 -2.80504168e-08] Intercept: 4504.3488372093025 Rounded coef: [560.242, 1019.463, -128.731, -86.153, 137.348, 55.212, -332.37, 376.363, 576.531, -144.129, -197.14, -0.0]
ridge = Ridge()
ridge.fit(X, y)
print("Ridge regularization coefficients/weights:\n", ridge.coef_)
print("\nIntercept:", ridge.intercept_)
print("\nRounded coef:", list(map(lambda c: round(c, 3) , ridge.coef_) ))
Ridge regularization coefficients/weights: [ 563.06457225 1018.94837879 -131.87332028 -86.746098 138.00511118 55.90311038 -332.3497885 386.45788919 566.34704706 -145.0713273 -99.25944108 -99.25944115] Intercept: 4504.3488372093025 Rounded coef: [563.065, 1018.948, -131.873, -86.746, 138.005, 55.903, -332.35, 386.458, 566.347, -145.071, -99.259, -99.259]
In contrast to L2 regularization, L1 resets the weights for some features.
Model coefficients as function of alpha
Let’s see how the weights change with an increase in the regularization coefficient alpha.
coefs_lasso is the matrix of weights of size: (number of regressors) x (number of features).
alphas = np.arange(1, 500, 50) coefs_lasso = np.zeros((alphas.shape[0], X.shape[1]))
For each coefficient value from alphas set we train the Lasso regressor and write the weights into the corresponding rows of the coefs_lasso matrix. Then we train the Ridge regressor and write the weights into coefs_ridge matrix.
print("Matrix `coefs_lasso` size:", (alphas.shape[0], X.shape[1]), "\nAlpha values:", alphas )
for i, al in enumerate(alphas):
lasso = Lasso(alpha = al)
lasso.fit(X, y)
for j, coef in enumerate(lasso.coef_):
coefs_lasso[i][j]=coef
#print(list(map(lambda c: round(c, 3) , coefs_lasso[i]) ))
# list(map(lambda c: round(c, 3) , coefs_lasso) ))
print("Lasso coefficients, head:\n", coefs_lasso[:5])
print("\nRounded Lasso coefficients:\n", list(map(lambda i: list(map(lambda j: round(j, 3), i)), coefs_lasso)))
Matrix `coefs_lasso` size: (10, 12) Alpha values: [ 1 51 101 151 201 251 301 351 401 451] Lasso coefficients, head: [[ 5.60241616e+02 1.01946349e+03 -1.28730627e+02 -8.61527813e+01 1.37347894e+02 5.52123706e+01 -3.32369857e+02 3.76363236e+02 5.76530794e+02 -1.44129155e+02 -1.97139689e+02 -2.80504168e-08] [ 4.10969632e+02 9.77019409e+02 -0.00000000e+00 -5.34489688e+01 9.19434374e+01 1.75372118e+01 -3.18125568e+02 3.22829934e+02 6.10031512e+02 -9.10689615e+01 -1.45066095e+02 -2.29877063e-08] [ 3.70077089e+02 9.35945490e+02 0.00000000e+00 -1.21619360e+01 4.88886342e+01 0.00000000e+00 -3.08805664e+02 2.69417263e+02 6.32502623e+02 -2.75042876e+01 -9.37749037e+01 -2.41652170e-08] [ 3.32835717e+02 8.91870058e+02 0.00000000e+00 -0.00000000e+00 0.00000000e+00 0.00000000e+00 -2.79616688e+02 2.11052030e+02 6.62920880e+02 -0.00000000e+00 -5.01551472e+01 -2.62761604e-08] [ 2.98134448e+02 8.45652857e+02 0.00000000e+00 -0.00000000e+00 0.00000000e+00 0.00000000e+00 -2.35571345e+02 1.24144807e+02 7.25379483e+02 -0.00000000e+00 -1.26461769e+01 -2.78781252e-08]] Rounded Lasso coefficients: [[560.242, 1019.463, -128.731, -86.153, 137.348, 55.212, -332.37, 376.363, 576.531, -144.129, -197.14, -0.0], \ [410.97, 977.019, -0.0, -53.449, 91.943, 17.537, -318.126, 322.83, 610.032, -91.069, -145.066, -0.0], \ [370.077, 935.945, 0.0, -12.162, 48.889, 0.0, -308.806, 269.417, 632.503, -27.504, -93.775, -0.0], \ [332.836, 891.87, 0.0, -0.0, 0.0, 0.0, -279.617, 211.052, 662.921, -0.0, -50.155, -0.0], \ [298.134, 845.653, 0.0, -0.0, 0.0, 0.0, -235.571, 124.145, 725.379, -0.0, -12.646, -0.0], \ [258.927, 799.237, 0.0, -0.0, 0.0, 0.0, -190.822, 72.076, 750.363, -0.0, -0.0, -0.0], \ [217.428, 752.721, 0.0, -0.0, 0.0, 0.0, -145.713, 37.715, 756.297, -0.0, -0.0, -0.0], \ [175.93, 706.204, 0.0, -0.0, 0.0, 0.0, -100.605, 3.661, 761.926, -0.0, -0.0, -0.0], \ [134.628, 659.633, 0.0, -0.0, 0.0, 0.0, -55.511, 0.0, 737.348, -0.0, -0.0, -0.0], \ [93.352, 613.055, 0.0, -0.0, 0.0, 0.0, -10.419, 0.0, 709.131, -0.0, -0.0, -0.0]]
coefs_ridge = np.zeros((alphas.shape[0], X.shape[1]))
for i, al in enumerate(alphas):
ridge = Ridge(alpha = al)
ridge.fit(X, y)
for j, coef in enumerate(ridge.coef_):
coefs_ridge[i][j]=coef
print(list(map(lambda c: round(c, 3) , coefs_ridge[i]) ))
[563.065, 1018.948, -131.873, -86.746, 138.005, 55.903, -332.35, 386.458, 566.347, -145.071, -99.259, -99.259] [461.179, 954.308, -41.565, -84.913, 126.604, 54.252, -313.275, 458.901, 481.444, -151.291, -101.627, -101.627] [403.977, 898.084, 5.674, -81.911, 117.941, 52.728, -298.409, 455.29, 467.431, -152.686, -102.102, -102.102] [366.604, 848.463, 34.027, -78.772, 110.68, 51.257, -286.125, 447.48, 455.754, -151.483, -102.005, -102.005] [339.745, 804.251, 52.49, -75.717, 104.403, 49.842, -275.486, 438.51, 444.764, -148.944, -101.586, -101.586] [319.159, 764.561, 65.152, -72.82, 98.879, 48.485, -266.003, 429.214, 434.235, -145.698, -100.965, -100.965] [302.636, 728.709, 74.138, -70.099, 93.956, 47.185, -257.391, 419.926, 424.122, -142.089, -100.209, -100.209] [288.913, 696.146, 80.66, -67.555, 89.529, 45.943, -249.471, 410.8, 414.409, -138.317, -99.361, -99.361] [277.213, 666.43, 85.459, -65.179, 85.519, 44.757, -242.124, 401.912, 405.086, -134.499, -98.449, -98.449] [267.031, 639.195, 89.012, -62.959, 81.865, 43.625, -235.264, 393.298, 396.138, -130.71, -97.493, -97.493]
The weights dependence on alpha dynamic
We visualize the dynamics of the weights as the regularization parameter alpha increases.
plt.figure(figsize=(10, 8))
for coef, feature in zip(coefs_lasso.T, df.columns):
plt.plot(alphas, coef, label=feature, color=np.random.rand(3))
plt.legend(loc="upper right", bbox_to_anchor=(1.4, 0.95))
plt.xlabel("Alpha")
plt.ylabel("Feature weight")
plt.title("Lasso")
plt.figure(figsize=(10, 8))
for coef, feature in zip(coefs_ridge.T, df.columns):
plt.plot(alphas, coef, label=feature, color=np.random.rand(3))
plt.legend(loc="upper right", bbox_to_anchor=(1.4, 0.95))
plt.xlabel("Alpha")
plt.ylabel("Feature weight")
plt.title("Ridge")
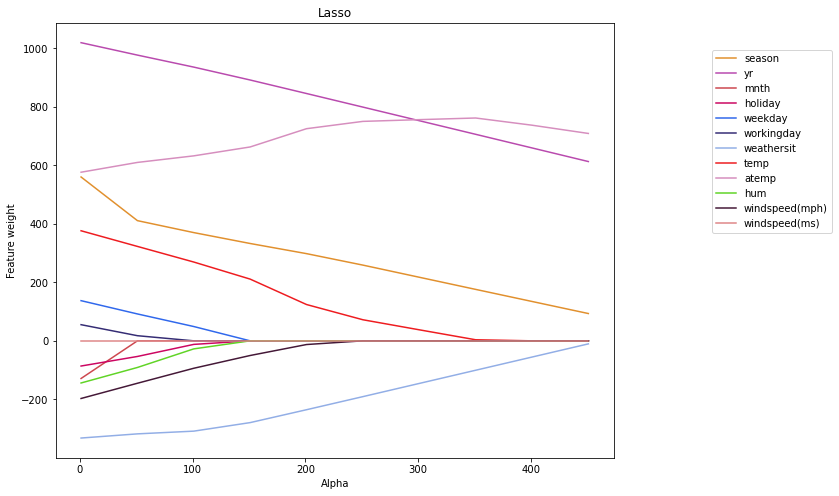
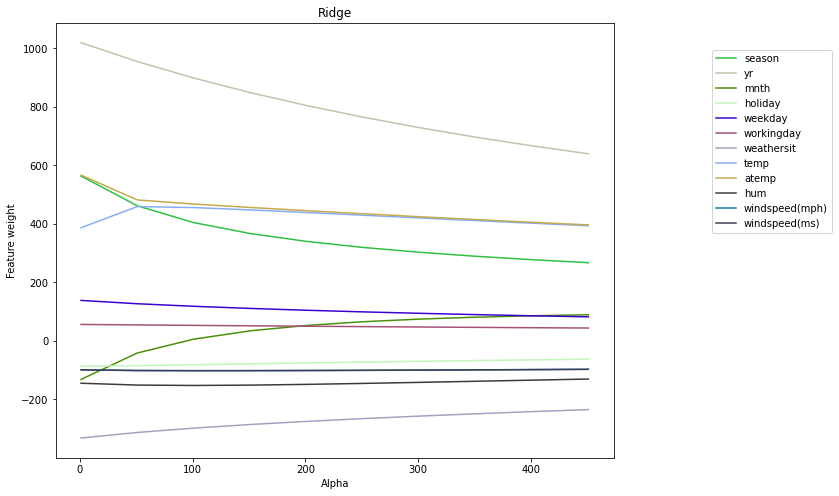
Sub-conclusions on Lasso and Ridge regularizators
We may do the following conclusions of the above figures:
- Lasso regularizator reduces weights more aggressively than Ridge at the same alpha.
- The regression model weights with the Lasso regularizator will tend/pursure to zero. The higher the alpha, the lower the complexity of the model. Then, for very large values of alpha, it is optimal to simply bring all the weights to zero.
- It is assumed that the regularizator excludes a feature if the feature coefficient is less than
0.001(<1e-3). At Lasso regularizator withalpha = 1, the weights for both “windspeeds” are not zero. But foralpha >= 50, the weights for both “windspeeds” are greatly reduced. As for the Ridge (L2) regularizator, the weights for collinear features depend little on alpha. Ridge does not exclude features when alpha increasing. - Lasso always excludes one of the duplicate attributes/features while Ridge only reduces the weight with one of them.
- Lasso regularizator is more fitting for selecting uninformative features.
We will work with Lasso (L1) regularizator to optimize regression model with regularization
We might see that when alpha changes, the model selects the feature coefficients differently. We need to choose the best alpha then.
We first choose a quality metric. We will take the optimized functional of the least squares method, i.e. Mean Square Error (MSE), as a metric. Now, how to calculate MSE for different alpha?
Cross-validation
- We can’t choose alpha by the MSE value on the whole training sample
- We neither can choose a single split of the sample into training and test sets (“holdout”), since we will tune the model in to specific “new” data, namely the test set.
- The best approach is cross-validation. We divide the sample into
Kparts, or blocks, and each time take one of them as a test, and make a training sample from the remaining blocks. We will make several partitions of the sample, try different alpha values on each one, and then average the MSE.
The cross-validation for regression in sklearn library is quite simple: there is a special regressor for this, namely LassoCV. It takes as input a list of alphas and calculates the MSE for each of them on cross-validation. After training (parameter cv=3), the regressor will contain the variable mse_path_, a matrix of size len(alpha) x K, where K = 3 (the number of blocks in cross-validation), containing the MSE values on the test for the corresponding runs. In addition, the variable alpha_ will store the selected (optimal) value of the regularization parameter, and in coef_, will have the trained weights corresponding to this alpha_.
Note that the regressor can change the order in which it iterates through the alphas values. In order to map it to the MSE matrix, it is better to use the regressor variable alphas_.
from sklearn.linear_model import LassoCV
alphas = np.arange(1, 100, 5)
reg = LassoCV(cv=3, random_state=0, alphas = alphas )
reg.fit(X, y)
print("Regularizator score (coefficient of determination\n R^2 of the prediction):", reg.score(X, y))
print("\nRegularizator MSE matrix head:\n", reg.mse_path_[:5])
print("\nRegularizator\"s `Alpha`:", reg.alpha_)
print("\nRegularizator coefficients:")
[(pair[0], round(pair[1], 4)) for pair in zip(df.columns[:-1], reg.coef_)]
Regularizator score (coefficient of determination
R^2 of the prediction): 0.800081404381094
Regularizator MSE matrix head:
[[863936.50981215 826364.11936907 862993.29751897]
[860479.31511364 821110.1817776 853075.13780626]
[857344.83606082 816153.27782428 843628.81286099]
[854526.73639431 811496.34805693 834654.45357263]
[852024.62341384 807139.39657173 826152.16399016]]
Regularizator`s `Alpha`: 6
Regularizator coefficients:
[("season", 532.019),
("yr", 1015.0602),
("mnth", -100.0395),
("holiday", -83.294),
("weekday", 132.5045),
("workingday", 51.5571),
("weathersit", -330.5599),
("temp", 370.6799),
("atemp", 581.3969),
("hum", -140.0074),
("windspeed(mph)", -191.7714),
("windspeed(ms)", -0.0)]
mean_mse_arr = np.array(reg.mse_path_).mean(axis=1)
#print(mean_mse_arr)
plt.figure(figsize=(10, 8))
plt.plot(reg.alphas_, mean_mse_arr, label=feature )
plt.xlabel("Alpha")
plt.ylabel("Mean MSE by fold")
plt.title("Lasso regularizator MSE")
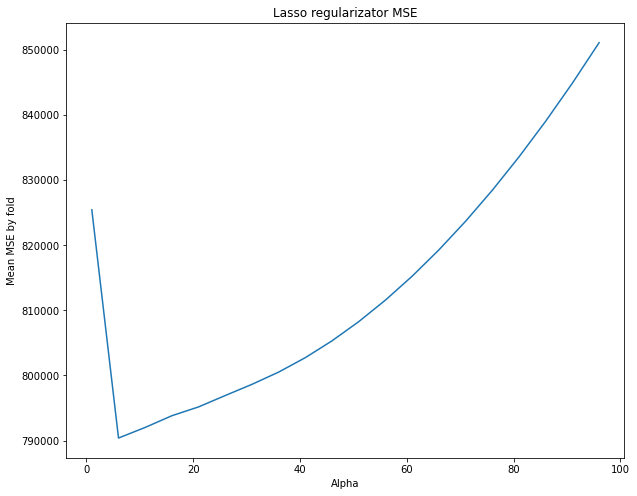
We have chosen a certain regularization parameter (alpha). Let’s look at what kind of alpha we would choose if we divided the dataset only once into training and test sets. We’ll consider the MSE graphs corresponding to single blocks (fold) of the dataset.
mse_columns = np.array(reg.mse_path_).transpose()
print("Minimum MSE for each column:", np.amin(mse_columns, axis=1), "\n" )
#print (reg.mse_path_)#[NumberOfColum])
for i, column in enumerate(mse_columns):
print("MSE for column {}:".format(i) , column.mean() )
plt.figure(figsize=(10, 8))
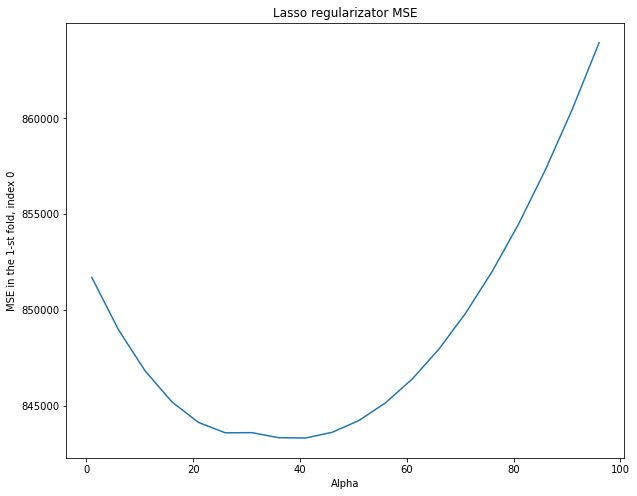
plt.plot(reg.alphas_, mse_columns[0], label=feature )
plt.xlabel("Alpha")
plt.ylabel("MSE in the 1-st fold, index 0")
plt.title("Lasso regularizator MSE")
Minimum MSE for each column: [843336.18149882 772598.49562777 745668.60596081] MSE for column 0: 848818.027469035 MSE for column 1: 797903.087624002 MSE for column 2: 794699.8496187156
fig, axs = plt.subplots(len(mse_columns))
fig.set_size_inches(12,12)
fig.suptitle("MSE in each of 3 Folds")
for i, col in enumerate(mse_columns):
axs[i].plot(reg.alphas_, col)
axs[i].set_xlabel("Alpha")
axs[i].set_ylabel("MSE, fold {}".format(i))
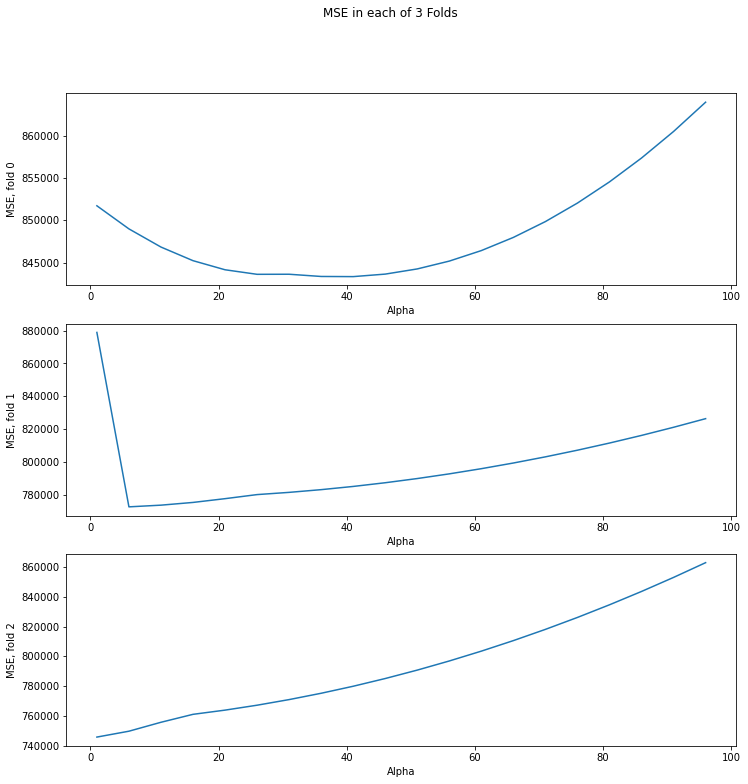
On each partition (fold), the optimal value of alpha is different. It might correspond to a large MSE on the other partitions. It turns out that we are tuning regularization parameter alpha in to specific training and control sets. When choosing alpha on cross-validation, we choose something “average” that will give an acceptable metric value on different dataset partitions.
Sub-conclusion on the Lasso regularization
- In the last trained model, there are 4 features with the highest (positive) coefficients: ‘season’, ‘yr’, ‘temp’, ‘atemp’.
- We may look at the visualizations of the `cnt` dependencies on these features that we drew in the “Dataset” block. There we may see increasing linear dependence of cnt on these features. It is logical to say (from common sense) that the greater the value of these feature weights, the more people will want to take bicycles.
- We select the features with the largest modulo negative coefficients: ‘weathersit’, ‘ windspeed(mph)’, ‘hum’.
- The graphs show a decreasing linear relationship with the target,
cnt. It is logical to say that the greater the magnitude of these feature weights, the fewer people will want to take bicycles. - The feature windspeed (ms) has coefficient close to zero (less than
1e-3). The linear model excluded it since it is a correlated (collinear) feature with windspeed(mph) from the model. One may see it at the graphs there. It is true that this feature does not affect the demand for bicycles since windspeed (ms) has already influenced it.
Conclusion
In the post we’ve built a linear model and evaluated the adequacy of the model. We’ve learned how to select features, and how to correctly select the regularization coefficient alpha (without tuning in the model to any particular piece of data).
When using the cross-validation we selected only a small number of folds/partitions (3). It’s because for each valid combination of them, we have to train the model several times (for different alpha) that is a time-consuming process esp. for large amounts of data.
